Recently, the increasing volume of data and application of neural networks have both forced to look at data format again. Previously, I thought the HDF5 format is the best for most of my application. The nice APIs to HDF5, e.g. H5py and DeepDish gives me both flexibility and easiness of using HDF5 to store and share my dataset. However, as my datasets start to grow substantially, loading them into the memory puts a significant burden on my I/O bus, especially I only need part of that dataset every time. Moreoever, the parallel processing the dataset by calling the same program multiples times with different parameters which may used different part of the dataset. Looking the entire dataset to the memory seems to be a waste of resource. This makes me to search for the alternative of HDF5 approach.
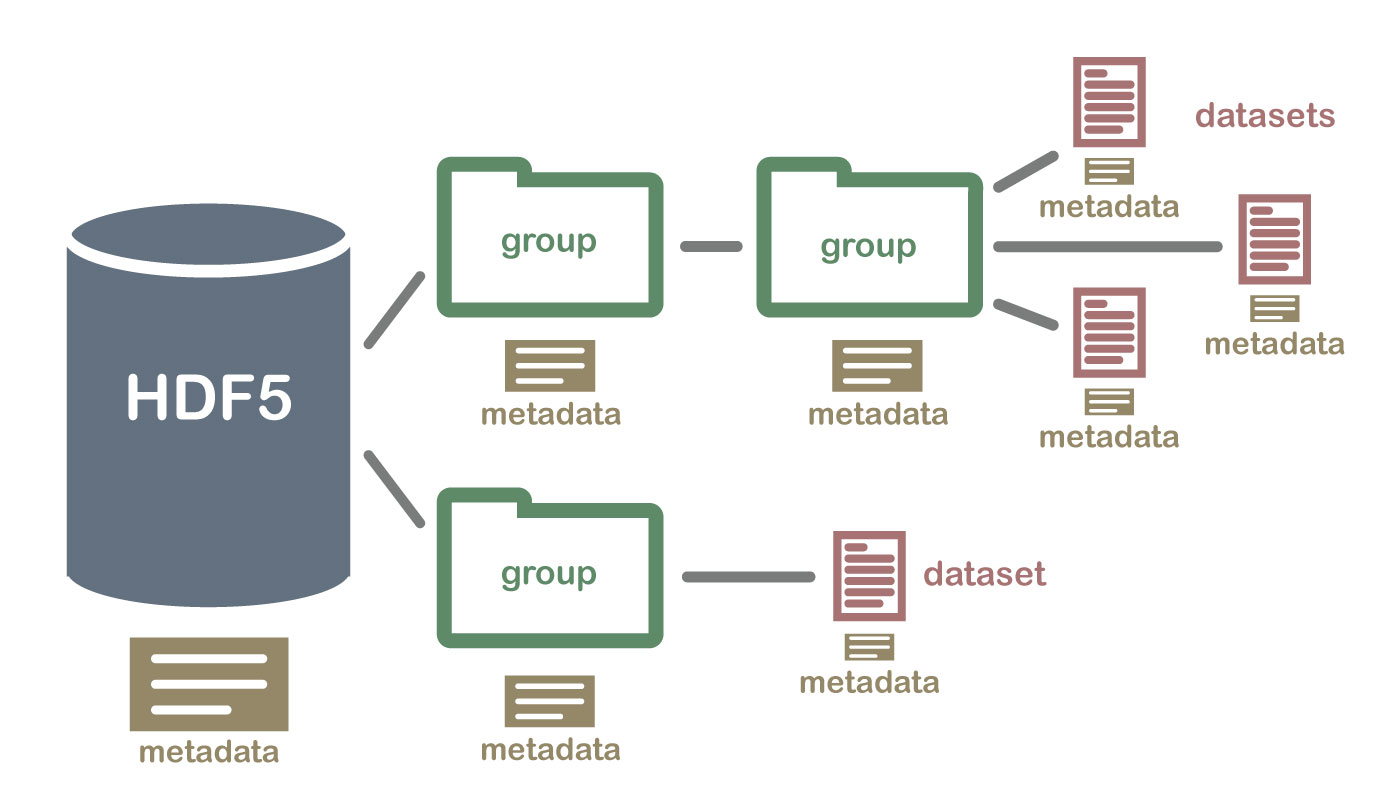
Dr. Cyrille Rossant’s recent post (and its follow-up) shares some similar problems as I encountered. The approaches he took is very interesting. As shown in the image above, HDF5 is close to have a mini-file-system inside the data format. This can be handled more efficiently by the file system. More importantly, the meta data written in plain text helps users to understand the structure and basics of the given dataset directly using OS without special tools. The question on stack overflow seems to resonate my point as well. If data sharing is needed, a simple tar/zip will easily package the entire folder into a single file.
However, the file format choice of the meta data is uncertain. The available options are json, yaml, and toml. I will continue the discussion in next post.